-
Introduction
-
Web Browser Requirements
-
Evidence System
-
What can users do in DGPD
-
Usage
- 1. Browse
- 2. Search
- 3. Download
- 4. Contact
-
Database Summary
Introduction
Dense granule proteins(GRAs) are secreted by the specific secretory organelle--dense granule of the Apicomplexans. Most of the GRAs locate within the parasitophorous vacuole (PV) where the parasite multiplies and maintain intracellular parasitism of nearly all nucleated host cells mainly by modifying the PV at the interface between the host cell and the parasite. The functions of these GRAs with different localization are also diverse, include participating in the formation of tubular membranes, regulating signaling pathways in host cells and affecting the transport of substances in the vacuolar membrane. To Integrate existing GRAs and analyze their functions are an necessary way for identifying novel GRAs. This work will help researchers to reveal the infection and pathogenicity mechanism of Apicomplexans.
Nearly all creatures can be the host of the apicomplexan species.In particular, Plasmodium falciparum and Toxoplasma gondii are the causative agents of two important human diseases: Malaria and toxoplasmosis, respectively. Neospora caninum causes neosporosis, one of the main causes of infectious abortion in cattle worldwide. Thus, apicomplexan parasites have a great influence on human health and animal husbandry, resulting in the low productivity and economic loss.
Over the past 20 years, our understanding of the exact biological mechanisms of GRAs has expanded dramatically. Unfortunately, this vast amount of knowledge has so far only concerned plasmodium and Toxoplasmosis. Although nine GRA proteins have been previously identified in N. caninum, including NcGRA1, NcGRA2, NcGRA6, etc. the exact biological function of these proteins is not fully understood. By the development of modern technologies, many studies reported that GRAs has potential applications in different aspects of functions. But little has been done to help the researchers in the research field of GRAs, thus we integrate rich GRAs data to develop a dedicated database, Dense Granule Protein Database (DGPD).
Web Browser Requirements
DGPD requires a modern web browser that supports JavaScript and cookies. To view complex details, you cannot block pop-up Windows. The following browsers have been thoroughly tested by the database monitoring system:
- Mozilla Firefox,Version 7 or above
- Internet Explorer,Version 11 or above
- Chrome,Version 8 or above
Evidence System
Levels of evidence for entries in DGPD.
| Level NO. | Category | Definition |
|---|---|---|
| Level 1 | Confirmed | The GRAs which has experimentally validated. |
| Level 2 | Likely | The highly suspected GRAs from main text or attachment of the articles. |
| Level 3 | Predicted | The homologous proteins of known dense granule proteins |
What can users do in DGPD
- Users can use keywords, genes and species data.
- Users can easily apply the search function that supports fuzzy queries to obtain content of interest in the field of the GRAs.
- The database monitoring system provides direct visualization of gene metadatas.
- Users can download standardized data from the database for free.
- Users can extend the database by submitting new data to us.
Usage
In the database monitoring system, we try to make it powerful and easy to use. This usage is intended for online services
Capitalized headings correspond to column headings in a web page table:
- Gene name: Gene accession number
- Evidence Level: Evaluation criteria for classifying data by source
- PMID: The index number used for literature search
- species: Toxoplasma, Neospora, Hammondia hammondi, Cystoisospora suis, Plasmodium falciparum
1. Browse
You can select one or more of the options listed in the browse area (species and Gene name).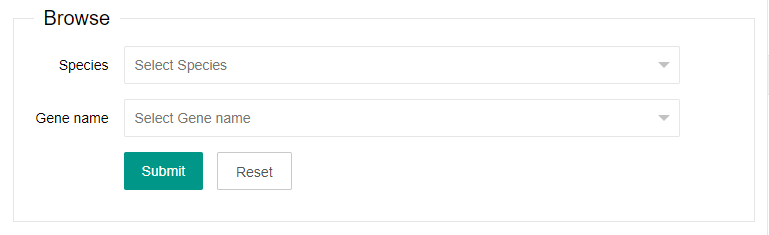
2. Search
You can enter a keyword to search in the database management system. The search field includes species name and gene name.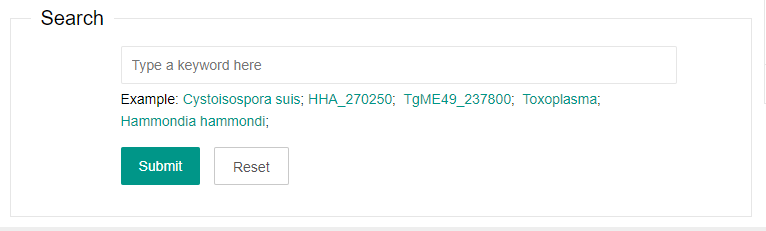
3. Download
We provide the option to download the full database. If you want to download, please select the menu "Download" and click the Download button.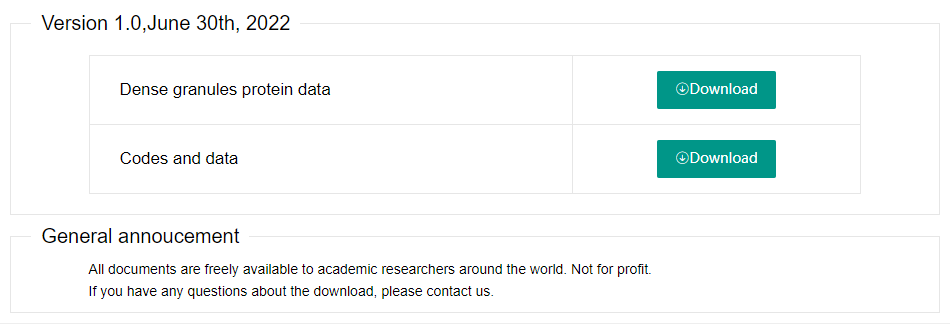
4. Contact
If you have any suggestions or questions not listed above, please contact us for details.(Email: hanghu@stu.ahau.edu.cn)
Database Summary (current version)
(A) DGPD statistics
(B) Type distribution
| leve 1 | leve 2 | level 3 |
| 110 | 37 | 98 |
| Species | Level 1 | Level 2 | Level 3 | Total | >
| Toxoplasma | 66 | 26 | 80 | 172 | >
| Hammondia hammondi | 11 | 0 | 18 | 29 |
| Plasmodium falciparum | 8 | 11 | 0 | 19 |
| Neospora | 16 | 0 | 0 | 16 |
| Cystoisospora suis | 9 | 0 | 0 | 9 |